Commits on Source (146)
Showing
- TeachingMaterials/2016/Antibodypedia.md 2 additions, 5 deletionsTeachingMaterials/2016/Antibodypedia.md
- TeachingMaterials/2016/EMBOSS_EBI.md 275 additions, 0 deletionsTeachingMaterials/2016/EMBOSS_EBI.md
- TeachingMaterials/2016/HPRexercise.md 55 additions, 0 deletionsTeachingMaterials/2016/HPRexercise.md
- TeachingMaterials/2016/ProteinBioinfo-MalvikaSharan.pdf 0 additions, 0 deletionsTeachingMaterials/2016/ProteinBioinfo-MalvikaSharan.pdf
- TeachingMaterials/2016/ProteinBioinfo-MalvikaSharan.pptx 0 additions, 0 deletionsTeachingMaterials/2016/ProteinBioinfo-MalvikaSharan.pptx
- TeachingMaterials/2016/UniProt.md 122 additions, 0 deletionsTeachingMaterials/2016/UniProt.md
- TeachingMaterials/2016/course-2016.md 68 additions, 0 deletionsTeachingMaterials/2016/course-2016.md
- TeachingMaterials/2016/course_content.md 10 additions, 8 deletionsTeachingMaterials/2016/course_content.md
- TeachingMaterials/2016/disprot.md 18 additions, 0 deletionsTeachingMaterials/2016/disprot.md
- TeachingMaterials/2016/elm.md 118 additions, 0 deletionsTeachingMaterials/2016/elm.md
- TeachingMaterials/2016/motif_visualization.md 507 additions, 0 deletionsTeachingMaterials/2016/motif_visualization.md
- TeachingMaterials/2016/multiple_sequence_alignment.md 81 additions, 0 deletionsTeachingMaterials/2016/multiple_sequence_alignment.md
- TeachingMaterials/2016/protein_database.md 65 additions, 0 deletionsTeachingMaterials/2016/protein_database.md
- TeachingMaterials/2016/sequence_similarity/images/BLOSUM62.png 0 additions, 0 deletions...ingMaterials/2016/sequence_similarity/images/BLOSUM62.png
- TeachingMaterials/2016/sequence_similarity/images/alignments.png 0 additions, 0 deletions...gMaterials/2016/sequence_similarity/images/alignments.png
- TeachingMaterials/2016/sequence_similarity/images/blosum.gif 0 additions, 0 deletionsTeachingMaterials/2016/sequence_similarity/images/blosum.gif
- TeachingMaterials/2016/sequence_similarity/images/descriptions.png 0 additions, 0 deletions...aterials/2016/sequence_similarity/images/descriptions.png
- TeachingMaterials/2016/sequence_similarity/images/graphic.png 0 additions, 0 deletions...hingMaterials/2016/sequence_similarity/images/graphic.png
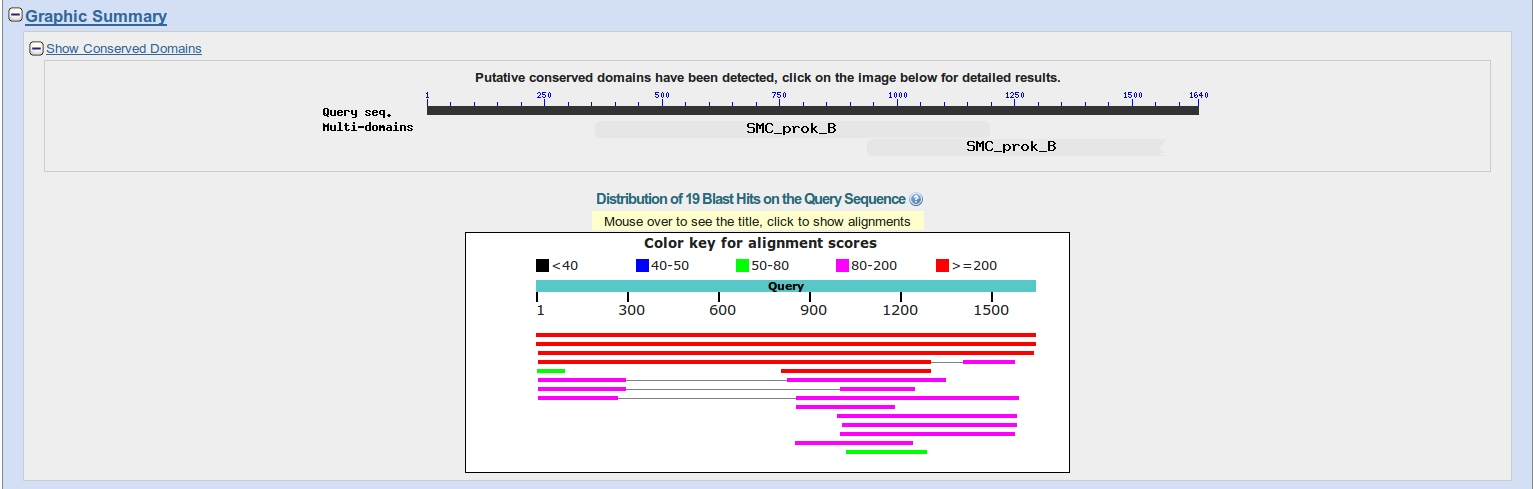
- TeachingMaterials/2016/sequence_similarity/images/graphicsummary.png 0 additions, 0 deletions...erials/2016/sequence_similarity/images/graphicsummary.png
- TeachingMaterials/2016/sequence_similarity/images/programselection.jpg 0 additions, 0 deletions...ials/2016/sequence_similarity/images/programselection.jpg
TeachingMaterials/2016/EMBOSS_EBI.md
0 → 100644
TeachingMaterials/2016/HPRexercise.md
0 → 100644
File added
File added
TeachingMaterials/2016/UniProt.md
0 → 100644
TeachingMaterials/2016/course-2016.md
0 → 100644
TeachingMaterials/2016/disprot.md
0 → 100644
TeachingMaterials/2016/elm.md
0 → 100644
This diff is collapsed.
TeachingMaterials/2016/protein_database.md
0 → 100644
44.8 KiB
180 KiB
28.2 KiB
126 KiB
104 KiB
34.1 KiB
62.1 KiB